# Upload & Storage
# Storage
Your storage is your personal virtual directory on the platform. It contains all the files you uploaded. Internally required conversions from your uploaded files may also be stored in your storage.
The list of uploaded files in your storage gathers:
- deployed images, ready to be inserted in a project;
- image uploads currently being converted;
- aborted image uploads due to conversion error, non-supported formats or any other unexpected error.
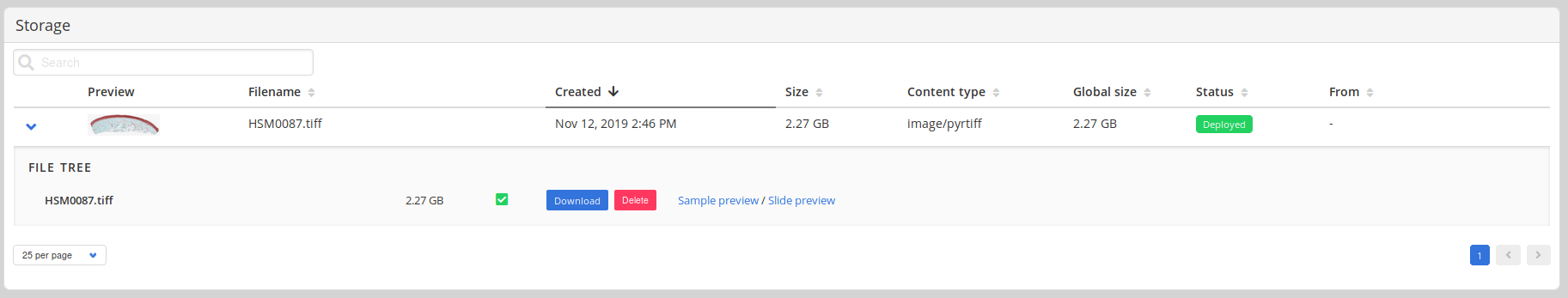
Files related to an image upload, such as internally required conversions can be shown in a file tree.
# Upload
To upload a file in Cytomine using the graphical interface, click on Add files and select the files to upload. By default, the files will be uploaded on your personal storage.
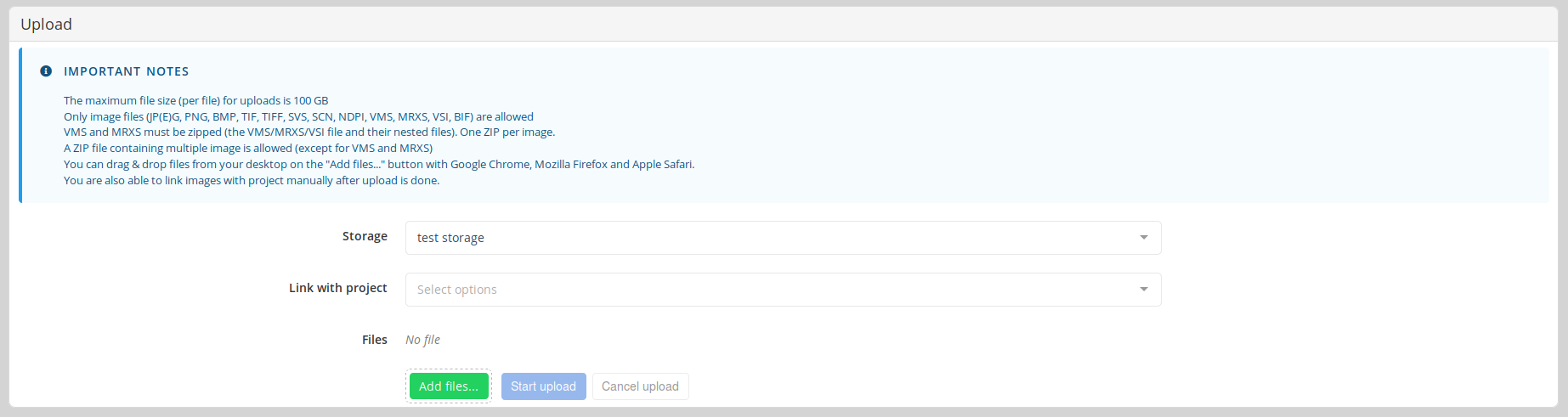
Optionally, it is possible to link the image uploads directly with a project. In any case, you can still associate them with one or several projects in the future, without uploading them again.
During the upload, a progress bar shows an indication of the file transfer to the Cytomine server.
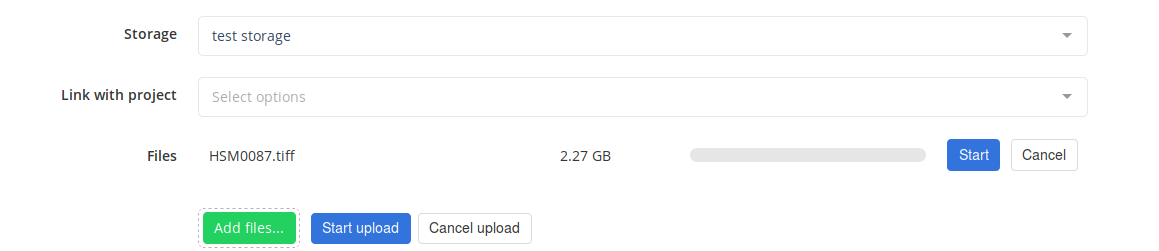
Once the upload is done, Cytomine tries to deploy your image on the server.
For native formats (the format is directly handled by Cytomine), the image is quickly available. Otherwise, Cytomine tries to convert the image to a natively supported format. This step can take some time, depending on the format and the size of the image. The current status of the file is displayed next to it.
# Uploaded file status
- UPLOADED: Files have been entirely received by Cytomine server, but they have not been treated yet.
- DETECTING FORMAT: Cytomine tries to detect the format of the image.
- EXTRACTING DATA: Cytomine is extracting data from some files (for example a directory or an archive).
- CONVERTING: Cytomine is converting the files to a native format.
- DEPLOYING: Cytomine is deploying your files in your storage (and optionally the projects).
If the process ended successfully, the uploaded file has one of these status:
- DEPLOYED: Data has been deployed successfully in your storage (and optionally the projects).
- CONVERTED: Data has been converted successfully and can be displayed in Cytomine.
- EXTRACTED: Data has been extracted from some files (for example a directory or an archive).
If the process has failed, the status gives you information about the issue:
- ERROR FORMAT: The format cannot be detected or is not supported.
- ERROR EXTRACTION: Data cannot be extracted from the file (for example from a directory or an archive).
- ERROR CONVERSION: Data cannot be converted.
- ERROR DEPLOYMENT: Unexpected error encountered during process.
# Supported files formats
In Cytomine, the format detection is not based on the file extension. Extensions given in the following table are purely informative.
Every pixel of an image has one or several sample(s). In greyscale images, only one sample is used. In RGB color images, 3 samples (Red, Green, Blue) are used. Each sample is encoded on a given amount of bits. 8-bit samples have values from 0 to 255 while 16-bit samples have values from 0 to 65535.
Some formats allow to store several channels (for example fluorescence) in the same file, some allow z-stacking and/or time series.
| Vendor / File format | Common extensions | Bit per sample | Sample per pixel | Channels | Z-Stack | Time | Remark |
|---|---|---|---|---|---|---|---|
| JPEG | .jpg, .jpeg | 8 | 1, 3 | - | - | - | |
| PNG | .png | 8, 16 | 1, 3 | - | - | - | |
| Planar TIFF | .tif, .tiff | 8, 16 | 1, 3 | - | - | - | |
| Pyramidal TIFF | .tif, .tiff | 8, 16 | 1, 3 | - | - | - | |
| JPEG2000 | .jp2 | 8 | 1, 3 | - | - | - | |
| DICOM | .dcm | 8, 16 | 1 | - | No | No | |
| Aperio / Leica SVS | .svs | 8 | 3 | No | No | ? | |
| Hamamatsu VMS | .vms | 8 | 3 | - | - | - | Files have to be grouped in a zip archive. |
| Hamamatsu NDPI | .ndpi | 8 | 3 | No | No | ? | |
| Leica SCN | .scn | 8 | 3 | No | No | ? | |
| 3DHistech MRXS | .mrxs | 8 | 3 | No | No | ? | Files have to be grouped in a zip archive. |
| Philips TIFF | .tif | 8 | 3 | - | - | - | |
| Ventana | .bif, .tif | 8 | 3 | ? | ? | ? | |
| Huron Technologies | .tif | 8 | 3 | - | - | - | |
| Cell Sens VSI | .vsi | ? | ? | ? | Files have to be grouped in a zip archive. | ||
| OME-TIFF | .ome.tiff | 8, 16 | 1, 3 | Yes | Yes | Yes | |
| Carl Zeiss CZI | .czi | 8, 16 | 1, 3 | Yes | Yes | Yes | |
| PGM | .pgm | 8, 16 | 1 | - | - | - | |
| BMP | .bmp | 8 | 3 | - | - | - | |
| GeoTIFF | .tif, .tiff | 8, 16 | 1, 3 | - | - | - | |
| GeoJP2 | .jp2 | 8 | 1, 3 | - | - | - | |
| Olympus Dot Slide | ? | ? | ? | Files have to be grouped in a zip archive. | |||
| MP4 | .mp4 | 8 | 1, 3 | - | - | Yes |
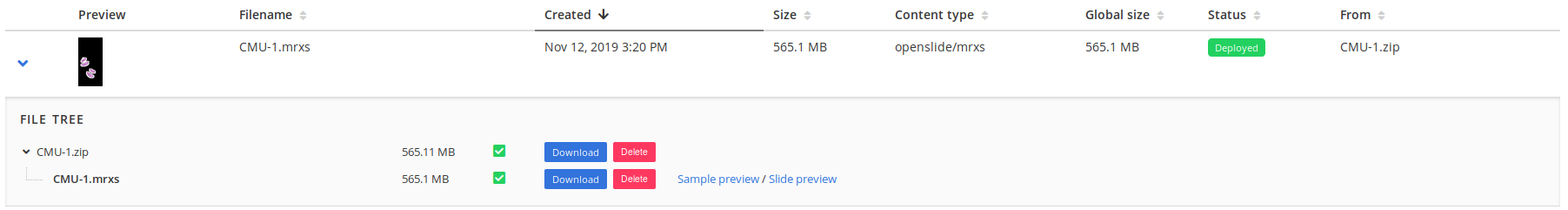
Hamamatsu VMS, 3DHistech MRXS and Cell Sens VSI formats are composed of multiple files.
Before uploading them, they have to be grouped into a zip archive.
For example, the MRXS file "CMU-1" that is available in the Cytomine cooperative open collection (opens new window) must have this structure:
- A
ziparchive calledCMU-1.zip. - A
mrxsfile calledCMU-1.mrxsat the root of the archive. - Other files (such as
datandinifiles) must be grouped into a folder next to the main file with the same name.
This example has this structure:
CMU-1.zip
|
--> CMU-1.mrxs
|
--> CMU-1/
|
---> Data0000.dat
|
---> Data0001.dat
|
---> ...
|
---> Data0022.dat
|
---> Index.dat
|
---> Slidedat.ini
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
# Known Issues
Potential Display Bug in Metadata for Certain Manufacturer WSI Images In our continuous efforts to keep our users informed about potential challenges that they may encounter, we would like to draw attention to a known issue that pertains to the display of metadata for WSI images from certain manufacturers.
Issue Overview: Under specific circumstances, users may notice an inconsistency or error in the metadata display for Whole Slide Images (WSI) provided by some manufacturers. This phenomenon seems to occur sporadically and may affect the usability of the meta data associated with these images.
Scenario: This issue typically arises when viewing metadata associated with some WSI images. While this anomaly does not always occur, it tends to be more frequent with specific WSI image providers.
Impact: The manifestation of this bug may result in the incorrect display of metadata for the affected WSI images.
Work around: None.
Status: As of the latest update, our development team is actively investigating this issue. We understand the inconvenience this might cause and appreciate your patience as we work towards a resolution. Updates will be provided in this section of the documentation as more information becomes available.
# Upload by batch
To import a large collection of images in Cytomine at once, you can group all your images in a zip archive and upload this archive.
