Cytomine Documentation portal
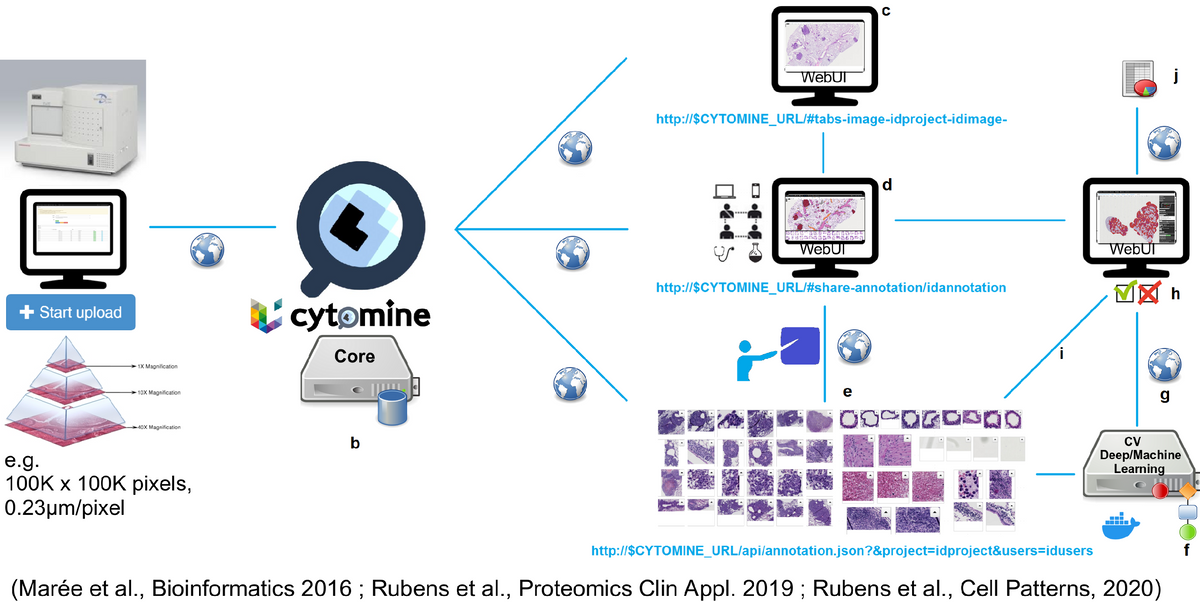
Cytomine software is a web platform for collaborative analysis of very large bio-medical images and semi-automatic processing of large image collections using machine learning algorithms. The software provides a very fast access to high-resolution images allowing users to annotate regions of interest and add semantic information to images and to annotations. Additionally, users have the possibility to run algorithms for semi-automated image analysis and share all their work with teammates using synchro- or asynchronous tools.
For imaging scientists and biomedical researchers
We are developing the Cytomine software for cutting-edge imaging modalities (beyond histology incl. 2D+c+z+t, hyperspectral data, spatial proteomics, imaging mass spectrometry, ...) and their combination using Image Groups and Annotation Links for multimodal studies.
For data and computer scientists
We are improving the Cytomine software execution architecture to support multiple types of algorithms and execution environments, and to perform benchmarking (see Biaflows project). We are also extending Cytomine internal data models and its RESTful API to support various types of data import/export.
For bioimage analysts and pathologists
We are developing novel AI algorithms to ease bioimage analysts and pathologists to analyze their data including interactive annotation tools and algorithms for various quantification tasks (cell counting, tumor delineation, content-based image retrieval,...). See ULiège R&D publications.
For other (non-biomedical) researchers
Through collaborations, our team is applying and extending some Cytomine features on large image sets from other fields such as digital collections of works of art, industrial quality control, remote sensing, ...
